-
Notifications
You must be signed in to change notification settings - Fork 24
Home
Facundo Muñoz edited this page Jun 6, 2020
·
28 revisions
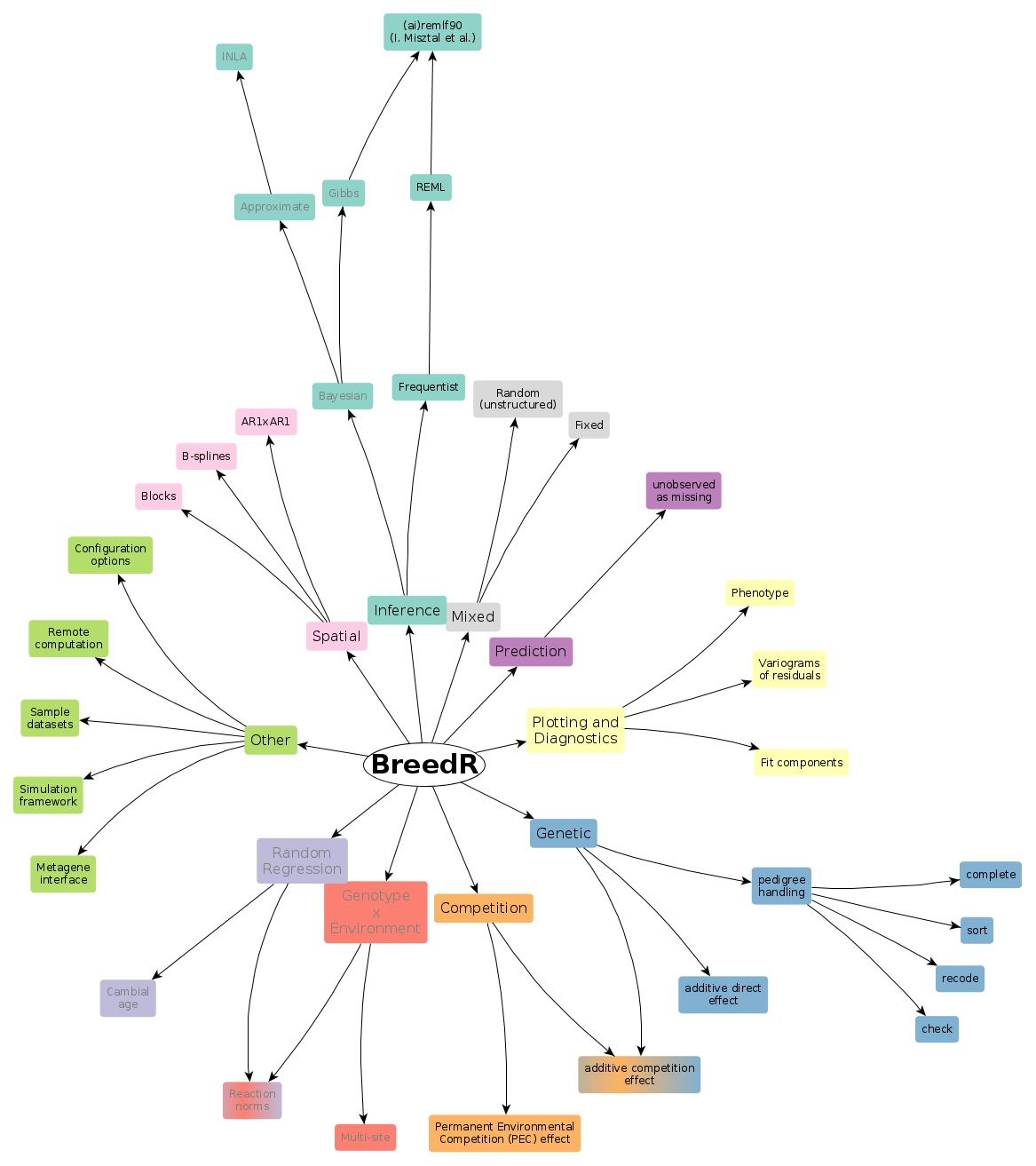
Check this presentation for a brief description of the package goals and modules.
The development on breedR has been halted until further notice. Hopefully, someone takes over the development or new funding is allocated to this project. I will be glad to help the developer to get started.
Questions? see the FAQ
The Getting started page provides simple guide to start using breedR for your own analyses.
- Single and multiple quantitative traits
- Handle pedigrees (check, recode, sort)
- Linear Mixed Models
- Additive genetic effects
- Spatial autocorrelation models
- Additive genetic competition model
- Permanent environmental competition model
- Simulation infrastructure
- Isotropic and anisotropic variograms of residuals
- Prediction of missing or hypothetical observations
- Real and simulated datasets and case-studies
- Reads metagene simulated datasets
More details:
- Check the latest additions in the NEWS file
- Check the available components and inference methods
- Random-regression models (see the UI specifications)
- Bayesian inference (Gibbs sampling)
- Genomic evaluation (GS3): for the moment, see the R package rgs3
We will build up a collection of datasets that are representative of most common scenarios in breeding and forest genetics. We will conduct beta testing using these datasets.
The scenarios, datasets and beta-testers will be organized through the Testing scenarios page.